Automatic Protein-Ligand interaction analysis
The new release of PLIP (Protein Ligand Interaction Profiler)
27 May 2021
The new release of PLIP, one of the most useful and complete tools to analyze protein-ligand interactions has been recently published in Nucleic Acids Research , where it was also published the original paper in 2015. The Protein-Ligand Interaction Profiler (PLIP) allows you easily analyze non-covalent interactions between biological macromolecules and their ligands based on PDB files. It can be used as a {web server](https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index) or locally. To use it locally it can be done through a containerized image or installing it via PyPy or from the source code (check the different options).
PLIP was originally developed by Schroeder’s group from the Biotechnology Center of the Technische Universität Dresden (https://tu-dresden.de). Since April 2020 PLIP is officially maintained by PharmAI GmbH. PharmAI states that “PLIP is probably the most wide spread open-source tool for detection of non-covalent interactions in biomolecules”, and I agree.
 Graphical abstract of [2021 PLIP publication](https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkab294/6266421#246639495)](https://raw.githubusercontent.com/MelchorSanchez/MelchorSanchez.github.io/master/static/img/blog/m_gkab294gra1.webp)
Graphical abstract of 2021 PLIP publication
I discovered PLIP less than a year after its original publication, around November 2016. Since then I have been regularly using it in almost a daily basis at work. There are some reasons because I like it. First of all it is quite precise. It is able to reproduce the reported binding modes of different structures. Then, it is code in python and open source available in GitHub. So you can read the code and understand it; you can learn what the code does. PLIP is not a black box. Thus as you understand the code, and as you can easily acces the code, you can tune it if needed. For instance you can custom the pymol representation or the different applied cutoffs. Now you can change the cutoffs, temporary, as a command line argument when executing the code (what is a really a nice addition), but in the initial versions you had to modify the config.py file (actually this is the way to permanently change it). For instance I used to change the BD_DIST (maximum distance to expand the considered binding site), the Hydrogen bond distance cutoff or the \pi stacking distance. PLIP determines interactions between ligand-protein residues based on distances and/or angles, depending on the measured interaction, so it’s important that you feel comfortable with the parameters. I recommend you to test PLIP at least in a simple benchmark to really understand its behaviour.
I also like PLIP because it generates nice outputd, useful and understanble. As an output you can get parsable files, in txt or xml format, png images of the protein-ligand interactions and also pymol sessions that then you can play with.
# Thresholds for detection (global variables)
BS_DIST = 10.0 #7.5 # Determines maximum distance to include binding site residues
AROMATIC_PLANARITY = 5.0 # Determines allowed deviation from planarity in aromatic rings
MIN_DIST = 0.5 # Minimum distance for all distance thresholds
# Some distance thresholds were extended (max. 1.0A) if too restrictive too account for low-quality structures
HYDROPH_DIST_MAX = 4.0 Distance cutoff for detection of hydrophobic contacts
HBOND_DIST_MAX = 3.5 #4.1 # Max. distance between hydrogen bond donor and acceptor (Hubbard & Haider, 2001) + 0.6 A
HBOND_DON_ANGLE_MIN = 100 # Min. angle at the hydrogen bond donor (Hubbard & Haider, 2001) + 10
PISTACK_DIST_MAX = 6.0 #5.5 # Max. distance for parallel or offset pistacking (McGaughey, 1998)
PISTACK_ANG_DEV = 30 # Max. Deviation from parallel or perpendicular orientation (in degrees)
PISTACK_OFFSET_MAX = 2.0 # Maximum offset of the two rings (corresponds to the radius of benzene + 0.5 A)
PICATION_DIST_MAX = 6.0 # Max. distance between charged atom and aromatic ring center (Gallivan and Dougherty, 1999)
SALTBRIDGE_DIST_MAX = 4.0 #5.5 # Max. distance between centers of charge for salt bridges (Barlow and Thornton, 1983) + 1.5
HALOGEN_DIST_MAX = 4.0 # Max. distance between oxy. and halogen (Halogen bonds in biological molecules., Auffinger)+0.5
HALOGEN_ACC_ANGLE = 120 # Optimal acceptor angle (Halogen bonds in biological molecules., Auffinger)
HALOGEN_DON_ANGLE = 165 # Optimal donor angle (Halogen bonds in biological molecules., Auffinger)
HALOGEN_ANGLE_DEV = 30 # Max. deviation from optimal angle
WATER_BRIDGE_MINDIST = 2.5 # Min. distance between water oxygen and polar atom (Jiang et al., 2005) -0.1
WATER_BRIDGE_MAXDIST = 4.1 # Max. distance between water oxygen and polar atom (Jiang et al., 2005) +0.5
WATER_BRIDGE_OMEGA_MIN = 71 # Min. angle between acceptor, water oxygen and donor hydrogen (Jiang et al., 2005) - 9
WATER_BRIDGE_OMEGA_MAX = 140 # Max. angle between acceptor, water oxygen and donor hydrogen (Jiang et al., 2005)
WATER_BRIDGE_THETA_MIN = 100 # Min. angle between water oxygen, donor hydrogen and donor atom (Jiang et al., 2005)
METAL_DIST_MAX = 3.0 # Max. distance between metal ion and interacting atom (Harding, 2001)
Config.py file form an old PLIP version (1.4.4) with thresholds modifiations
From version 1.4.4/1.4.5 (the latest version until pharmAI started to maintain the code) to version 2.1.3 (july 2020) and to version 2.2.0 (actual) several changes have been performed, as can be seen in the changelog. The evolution is specially notable if you compare with the first versions. Now PLIP is an even stronger interaction profiler that can handle protein-ligand interactions. But not only that, because now protein-peptide interactions, DNA/RNA-ligand interactions and interactions within one chain can be also detected. Moreover you can analyze different complexes in batch mode (at the beginning you need to script a loop of executions over different complexes). That is really useful for analyze, for instance, Virtual Screening, docking, results.
Regarding multiple complexes analysis, probably what I missed now is the possibility of upload a full MD trajectory file. However there are ways to handle it like upload the trajectory with mdtraj, pytraj or parmed, for example, extract PDB files from the different frames and then analyze them with PLIP (a very simple example can be found here). Other thing I would change is that the parsable txt or xml files you can get as output, could be somehow csv files. This will simplify the automatization of the extraction/analysis of the diverse interactions. Is true that it is not something too much complicated with the actual format but a csv file would be even easier to parse using Pandas, for example. And an automatic parsing is something you will need to do at some point. For few PDB files is fine to look at the report files, but if you want to go further you need to automatize the process. For instance to filter out the compounds that perform a specific interaction from a virtual screening (docking based) of a database with thousands of compounds or if you want to check during how many frames in an MD simulation a certain interaction is established, you need to automatize the interactions extraction and filtering. And to do that you need to create an script to automatize files parsing. However is true that, if well conceived, you only need to do that once (a possible example can be found here).
traj=pt.iterload('/PATH/to/the/MD/target_ligand_solvent-w-MD.dcd', \
top='/PATH/to/the/topology/file/\
target_ligand_solvent.prmtop', frame_slice=[(0, 100, 10),])
traj=traj.strip(':WAT,:Na+')
count=0
for frame in traj:
count=count+1
pt.write_traj('trajectory_frame'+str(count)+'.pdb', traj=traj, overwrite=True)
pt.write_traj('trajectory.pdb', traj=traj, overwrite=True)
plip=('plipcmd -f ' + trajectory_frame'+str(count)+'.pdb -t')
run_command(plip)
Analyze interactions of diverse frames from a NAMD trajectory with mdtraj and PLIP. Full sctipt can be found here
PyMol representation of extracted frame1 from the parsed trajectory
tree = ET.parse(plipoutput)
root = tree.getroot()
number_detected_ligands=len(root.findall("bindingsite"))
ligands_name=[]
ligtype=[]
longname=[]
for i in range(len(root.findall("./bindingsite/identifiers/"))):
if root.findall("./bindingsite/identifiers/")[i].tag == 'hetid':
ligands_name.append(root.findall("./bindingsite/identifiers/")[i].text)
if root.findall("./bindingsite/identifiers/")[i].tag == 'longname':
longname.append(root.findall("./bindingsite/identifiers/")[i].text)
if root.findall("./bindingsite/identifiers/")[i].tag == 'ligtype':
ligtype.append(root.findall("./bindingsite/identifiers/")[i].text)
Example code for parsing a PLIP output txt file. Full sctipt can be found here
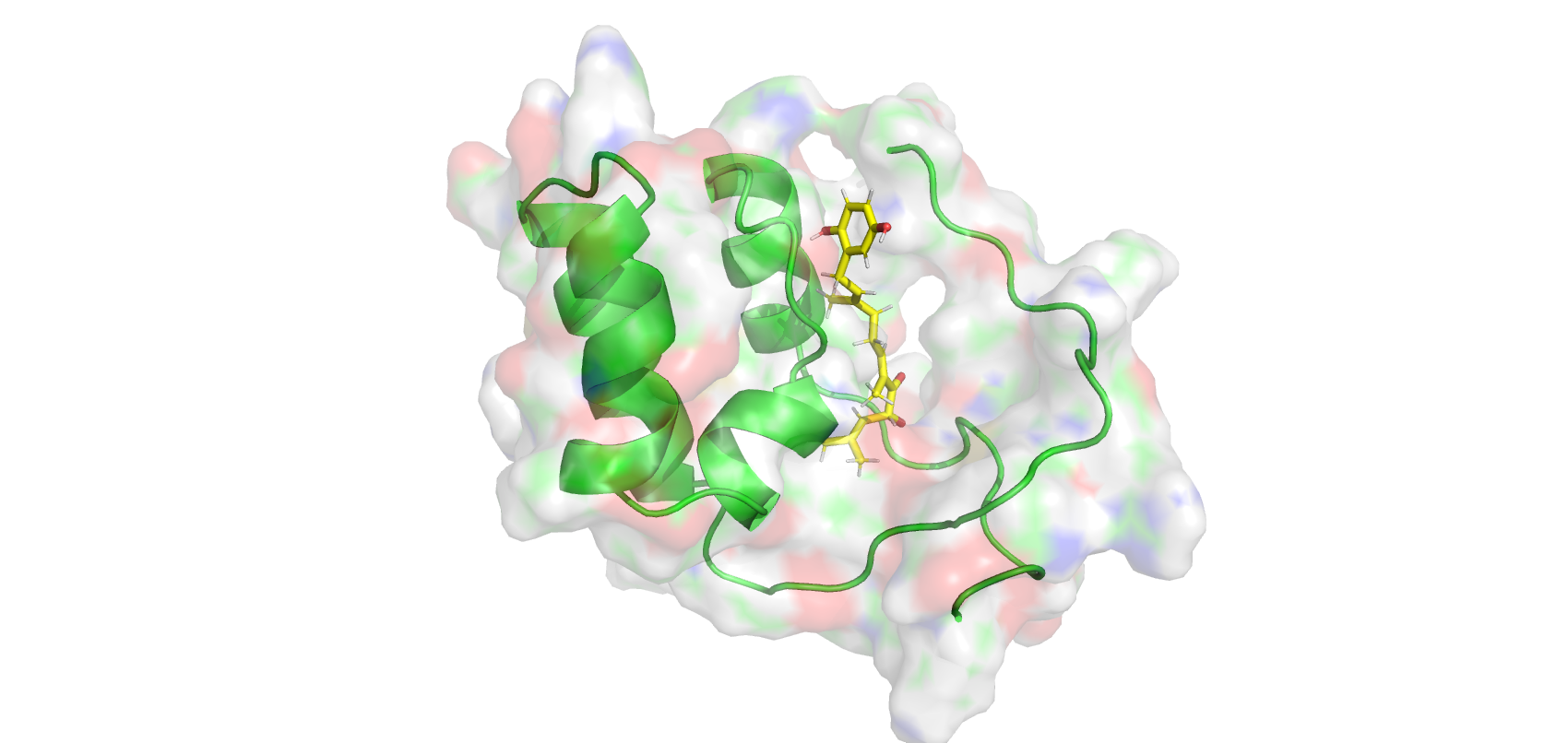
. A protein that I widely studied during my [PhD](https://www.tesisenred.net/bitstream/handle/10803/288044/MSM_PhD_THESIS.pdf?sequence=6&isAllowed=y) PLIP PyMol output of ANP ligand interactions with [1GS5](https://www.rcsb.org/structure/1gs5). A protein that I widely studied during my [PhD](https://www.tesisenred.net/bitstream/handle/10803/288044/MSM_PhD_THESIS.pdf?sequence=6&isAllowed=y)](https://github.com/MelchorSanchez/blog_post_example_scripts/blob/main/_associated_files/plip_parsing_example/1gs5_plip_output.png?raw=true)
PLIP PyMol output of ANP ligand interactions with 1GS5.. A protein that I widely studied during my PhD
For me PLIP is a must for a bioinfomatician/chemoinformatician/computational biologist/computational chemist (in another post we could talk about why the same thing can have a different name depending on your background). There are other protein-ligand interaction profilers out there, some of them with the capacity of generating 2D maps like the ones from Schrödinger suite, Discovery studio , Inteligand or also the open source LigPlot(+). But, in my humble opinion, none of them offers the flexibility of PLIP, as a consequence of be an open source python-based tool, and are not as complete as PLIP due to the high number of different interactions it can detects in different scenarios.
PD-1: Automatization is fine, is a goal to reach in almost any step of a Drug Discovery pipeline. To detect protein-ligand interactions, it helps a lot, specially while working with thousands of complexes. However it shouldn’t be forget that human knowledge is essential; take a look to the most promising compounds based on an interaction profiling is highly recommended. As I read in a recent Linkedin post “Visual exploration: What really makes the difference between docking analysis and throughout evaluation when looking for a hit molecule”. Tools like PLIP alleviate the checking necessity, but can’t avoid it.
PD-2: For a full documentation of running options and output formats of PLIP, please, refer to the Documentation.